A tutorial for microbial genome-scale metabolic modelling: [1] using models in MATLAB
This is my first post of a series of tutorials for constructing genome-scale metabolic models (GEMs) for single-cellular microbes. In this tutorial, I follow the protocol created by Heirendt et al. to demonstrate reading and visualisation of existing GEMs1.
1. Materials
- Windows 10 with git (or GitHub Desktop) installed.
- MATLAB R2019a. Thanks to the University of Melbourne for providing her students a licence to access this software.
- COBRA Toolbox v3.0 (GitHub repository, installation guide). The protocol paper publishing this toolbox provides a detailed user manual1.
- GEM iCM925 for a butanol-producing strain Clostridium beijerinckii NCIMB 80522 and iHN637 for C. ljungdahlii DSM 135285. See my last post for more details of this model. In addition, OpenCOBRA offers a repository of GEMs for public studies.
Installation
Download code through PowerShell
cd C:\Matlab\toolbox
git clone --depth=1 https://github.com/opencobra/cobratoolbox.git cobratoolbox
Then in MATLAB
cd C:\Matlab\toolbox\cobratoolbox % the installation folder of COBRA Toolbox
initCobraToolbox % It takes quite a while to finish.
testAll % It takes more than an hour to run a test suite locally.
I saw an error of an undefined function or variable rdir in the first time of initialisation. This problem was gone after restarting and reinitialising the toolbox. So eventually, my “COBRA Toolbox is up-to-date”.
Initialisation
According to the protocol for COBRA v3.01, we can create a startup file under the working directory.
userpath('C:\Analysis\GSM_demo') % specify the default user work folder
cd C:\Analysis\GSM_demo
edit startup.m % create the file under the work folder. Add "initCobraToolbox" in it.
MATLAB will spend some time to automatically load and initialise COBRA every time you run it.
2. Reading and writing genome-scale metabolic models
I load the model of C. beijerinckii NCIMB 8052 in the SBML format in this demonstration.
pwd % MATLAB should be working at the folder you specified using the userpath command.
model1 = readCbModel('iCM925.xml') % This command also prints the model description.
model2 = readCbModel('iHN637.xml')
Note that the filename of the model must be a character array. Hence a string such as “iCM925.XML” causes an error message. This syntax differs from R.
It is straightforward to write a model using COBRA Toolbox.
writeCbModel(model1, 'iCM925_Cb.xml') % Filename extension determines the output format.
> In writeSBML (line 175)
In writeCbModel (line 218)
Warning: Metabolite pgp_CB[C_c] has formula C38.7H72.8O13P2. FBC 2.1 only allows integer values for coefficients.
Discarding the formula. % A few warnings displayed for this model.
writeCbModel(model2, 'iHN637_Cb.xml')
The text layout of iCM925_Cb.xml looked much better than iCM925.xml (which does not have any newline characters) after writing the model using the toolbox. Notably, no error or warning message appeared when MATLAB was saving model2 — everything went quite neatly, suggesting that the first model is not compatible with COBRA Toolbox.
3. Displaying a model
We need tools other than MATLAB to visualise GEMs as metabolic networks.
Cytoscape v3.7 with cy3sbml plugin
The cy3sbml plugin is a generalist tool for visualising GEMs3, 4. The plugin and its installation guide can be freely accessed in the Cytoscape App Store. Basically, the plugin can be installed through the App Manager of Cytoscape. Hence there is no need to download a stand-alone installer from the website. I installed the version 0.2.7 to show the model iCM925 for this post.
File > Import > Network from File (It often takes a while to load the model as a network)
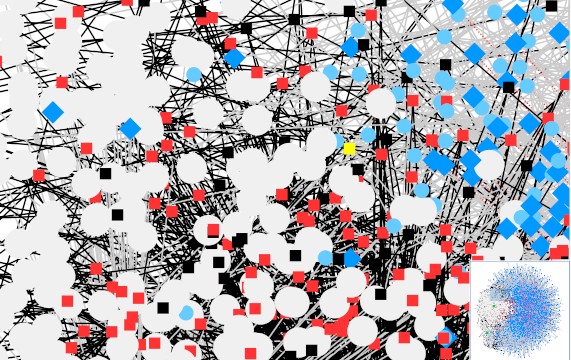
Figure 1: a Cytoscape view of the model iCM925.
As we can see in Figure 1, the Cytoscape view is way too complicated to be meaningful. However, we could display a subnetwork by choosing the nodes of interest and only show relevant links. In summary, there are five kinds of nodes in the model iCM925:
- Compartment, such as Cytoplasm, Mitochondria and Extracellular in model iCM925. Compartments are high-degree hexagon nodes.
- (Chemical) Species, each has a compartment attribute showing the node ID (e.g., c_e) of the compartment that it belongs to. Shape: large filled circles.
- Reaction, which links to kineticLaw (ID). Shape: little squares. Colours: black (reversible), red (irreversible).
- KineticLaw, which links to reactions via the attribute of reaction IDs. Shape: small filled circles.
- LocalParameter of reactions (linked through reaction IDs). Shape: diamond.
According to the node classes, links in the model include species_compartment, localParameter_kineticLaw, reaction-reactant, reaction-product and so forth.
In addition, the model is comprised of three networks: All, Base and Kinetic.
ModelExplorer
Recently, the Almaas lab from Norway published an alternative tool for visualisation, investigation, edition and correction of GEMs — the ModelExplorer v2.06. It runs on either Linux or Windows and can be started through a command line or graphical user interface (GUI). It is evident in Figure 2 that this tool has improved the network visualisation compared to cy3sbml.
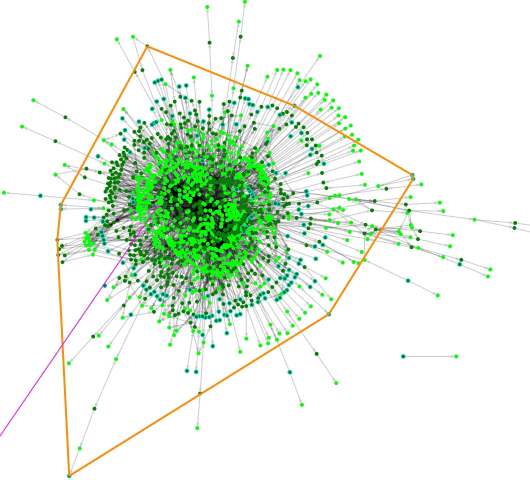
Figure 2. a ModelExplorer view of model iHN637. The orange polygon denotes chemical species (metabolites) within the compartment cytosol. The other compartment is extracellular space, which is not enclosed by any polygon. Note that a few metabolites belonging to the extracellular space are actually placed within the polygon denoting the cytosol by ModelExplorer for the optimal layout. The purple line indicates a reaction I was searching for.
As a norm of open-source bioinformatic software, ModelExplorer does not offer a user-friendly GUI. For example, you cannot paste the path of your model when you try to open it. As a result, you may want to put your model under the software’s folder to save time. Moreover, there is a trick to show or edit node information — you need to click your right mouse button (RMB) in an empty space to recover the dynamic view of the text panel as you hover over nodes, or the panel freezes after your first RMB click.
Despite ModelExplorer shows an evident advantage over the Cytoscape + cy3sbml approach, it failed to open the model iCM925 and reported a number of warnings. This problem was partly fixed (iCM925_Cb.xml) through rewriting the model using the COBRA Toolbox (see Section 2) — the new model file can be eventually visualised by ModelExplorer. However, the compartment names were lost. So there must be something wrong in the format of the original model which was published eight years ago.
.\ModelExplorer2\ModelExplorer.exe iCM925.xml
...
Warning! In FBA_model::load could not load reactant stoichiometry for R_ex_thm_e! Assumed = 1.0.
Warning! In FBA_model::load could not load product stoichiometry for R_ex_thm_e! Assumed = 1.0.
/////////////////////////////////////////////////////////////////////////////////////////
// //
// Opened Model: Flux Unit: Species Number: Reaction Number: //
// C__beijerinckii__iCM926 mmol_per_gDW_per_hr 900 938 //
//
/////////////////////////////////////////////////////////////////////////////////////////
References
- Heirendt, L. et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 14, 639–702 (2019).
- Milne, C. B. et al. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst. Biol. 5, 130 (2011).
- Martyushenko, N. & Almaas, E. ModelExplorer - software for visual inspection and inconsistency correction of genome-scale metabolic reconstructions. BMC Bioinformatics 20, 56 (2019).
- König, M., Dräger, A. & Holzhütter, H.-G. CySBML: a Cytoscape plugin for SBML. Bioinformatics 28, 2402–2403 (2012).
- Nagarajan, H. et al. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Fact. 12, 118 (2013).
- Martyushenko, N. & Almaas, E. ModelExplorer - software for visual inspection and inconsistency correction of genome-scale metabolic reconstructions. BMC Bioinformatics 20, 56 (2019).