A tutorial for microbial genome-scale metabolic modelling: [2] building a draft metabolic network from genome annotations of a single bacterial isolate
In my first post of this tutorial, I have demonstrated basic ways to inspect a genome-scale metabolic model (GEM). Now, let’s get our hands dirty — to reconstruct a draft metabolic network from genome annotations, which is the starting point of the protocol proposed by Thiele and Palsson for bottom-up GEM construction1, 2. In this post, we will be using several bioinformatic tools to reconstruct draft metabolic networks from annotations of the fully resolved chromosomal genome of Clostridium beijerinckii str. NCIMB 8052 (KEGG organism code: cbe; PATRIC genome ID: 290402.41), a well-known butanol-producing microorganism. A GEM iCM925 of this strain has been published by Milne et al3.
1. AutoKEGGRec
This MATLAB function, published by the Almaas Lab in 2018, is probably the easiest and fastest way for reconstructing a metabolic network from genome annotations2. Moreover, it supports simultaneous network reconstructions for an array of bacterial isolates. Nevertheless, it worth noting that this tool is restricted to published complete genomes in the KEGG database.
1.1. Installation
Simply download AutoKEGGRec.m from the Almaas Lab’s website and copy it to a MATLAB search path (run command pathtool to show all paths). No other steps are required.
The code is also publicly available on GitHub.
Adding a folder into the list of search paths
Run pathtool and choose the correct folder by clicking the Add Folder button in the Set Path dialogue box.
1.2. Network reconstruction
Given a KEGG Org code, we only need a single command line to carry out this
outStruct = AutoKEGGRec("cbe", 'SingleRecs', 'writeSBML'); % one network per organism
Troubleshooting
Error using toc
You must call TIC without an output argument before calling TOC without an input argument.
Error in AutoKEGGRec (line 268)
time = toc;
Solution: add a tic command before AutoKEGGRec, then the job finished successfully.
tic;
outStruct = AutoKEGGRec("cbe", 'SingleRecs');
% Actually, I should have used AutoKEGGRec("cbe", 'SingleRecs', 'writeSBML').
% However, I decided to test the speed of network reconstruction without being affected by the I/O speed of my hard driver.
% So I removed the option of writing an SBML file.
The selected flags:
SingleRecsflag
Parpool could not be started: it is unavailable or already running.
KEGG organism codes:
cbe
Starting accessing the KEGG Database for the requested Organisms.
All organism URLs have been built, and the reactions, compound and EC number linkage have been downloaded.
Time so far running AutoKEGGRec: 474 seconds.
All organism gene-to-reaction linkages have been downloaded.
Compound-to-reaction and reaction-to-enzyme matrices built. Starting organism-to-reaction matrix now.
Matrices built.
Commencing assembly of reconstruction(s).
Number of KEGG reactions for the KEGG organism(s) queried: 1303
Downloading all reactions from KEGG (this takes a while).
Progress:
....................................................................................................
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
All reactions needed have been fetched.
Time so far running AutoKEGGRec: 2778 seconds.
Number of unque KEGG compounds for the organism(s) queried: 1286
Downloading all compounds from KEGG (this takes a while).
Progress:
....................................................................................................
||||||||||||||||||||||||||||||||||||||||
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
All compounds needed have been fetched.
Time so far running AutoKEGGRec: 5043 seconds.
Adding KEGG annotations to the compounds.
Building consolidated reconstruction.
Adding KEGG annotations to the reactions.
Adding protein annotations and updating gene names for each organism:
Organism: cbe
Building single reconstructions
Identifying major components and finding percentage of reactions present in them.
Reactions in major component of the cbe reconstruction: 99.01%
AutoKEGGRec summary:
AutoKEGGRec completed its instructions in 5078 seconds.
Organisms received as input: 1
KEGG reactions assessed: 1303
Reactions marked as redundant: 27
Reactions marked as moved: 49
Reactions containing glycans: 6
Reactions involving polymerization: 8
Reactions producing their own substrate: 30
Unique compounds retrieved from KEGG: 1286
Generic (massless) compounds identified: 323
Reactions omitted for containing generic compounds: 280
Single-organism reconstructions created for all the input organisms.
Thanks for using AutoKEGGRec. Please cite our paper:
E.Karlsen, C.Schulz and E.Almaas
Automated generation of genome-scale metabolic draft reconstructions based on KEGG
BMC Bioinformatics 2018
The function was run within MATLAB R2019a on a HP Pavilion 15 laptop with a 8 GB RAM and an AMD Quad-Core Processor A6-5200 (2.0GHz, 2MB L2 Cache). The runtime is comparable to that listed in Table 2 of the article by Karlsen, Schulz and Almaas2.
1.3. Saving the resulting network as an SBML file
save('cbe_draft_20190605.mat', 'outStruct'); % save the workspace variable outStruct
% save network data within the struct array
net = getfield(outStruct, 'singleStructs');
writeCbModel(getfield(net, 'cbe'), 'cbe.xml');
Tip: double-clicking outStruct in the Workspace tab of MATLAB shows fields within this struct variable.
1.4. Network visualisation
The resulting network cbe.xml can be displayed using ModelExplorer4 (Fig. 1), also developed by the Almaas Lab.
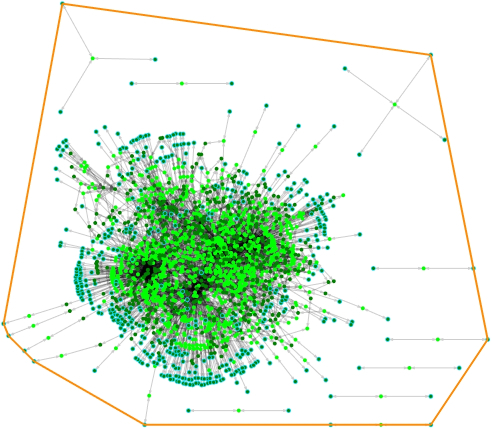
Figure 1. The draft metabolic network constructed using AutoKEGGRec for C. beijerinckii NCIMB 8052. The only compartment here is Cytoplasm.
2. ModelSEED-like pipeline from PATRIC
ModelSEED is a sophisticated web-based pipeline for generating draft GEMs from RAST genome annotations5. PATRIC (Pathosystems Resource Integration Center) implements a pipeline primarily based on the ModelSEED framework6, 7.
 **Figure 2. The pipeline of ModelSEED for reconstructing GEMs.** This figure comes from the original software [article](https://www.nature.com/articles/nbt.1672)5.
**Figure 2. The pipeline of ModelSEED for reconstructing GEMs.** This figure comes from the original software [article](https://www.nature.com/articles/nbt.1672)5.
2.1. Model reconstruction
It is simple to build a draft model (accessed on 6 June 2019) using ModelSEED. However, since options PATRIC Microbes or RAST Microbes under ModelSEED > Build Model on the website modelseed.org did not work for me (using Firefox, Chrome or Microsoft IE), I use the PATRIC > SERVICES > Metabolomics > Model Reconstruction instead. The genome ID 290402.41 of C. beijerinckii NCIMB 8052 was used for Select a Genome. I chose a complete media for model reconstruction and submitted the job by clicking the “Reconstruct” button. The webpage returned a message saying “Model Reconstruction Job has been queued.” The job can be found under WORKSPACES > My Jobs.
2.2. Result inspection
Results can be found under the user-specified folder when the Job Status becomes completed. In my case, seven output files were found under the subdirectory Cbe8052. The model file Cbe8052.sbml amongst these files was downloaded and visualised with ModelExplorer. The webpage for Cbe8052 also displayed a summary of the model: 918 genes, 1388 compounds, 1239 reactions and one biomasses.
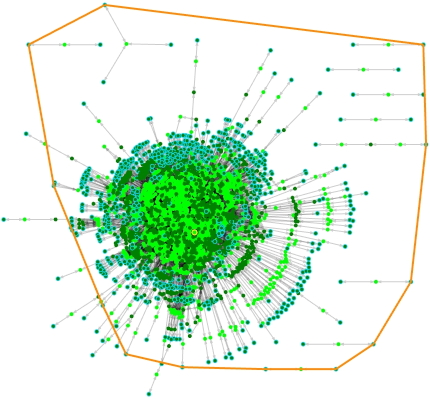
Figure 3. The draft model produced on PATRIC. The only compartment is Cytosol_0.
3. MATLAB + RAVEN v2.0
As I have introduced in a previous post, RAVEN is a widely used tool for GEM reconstruction. The GitHub wiki of this package provides an installation guide. A series of detailed tutorials are offered by authors as well, making RAVEN an appropriate starting point for beginners. As such, I am not going to repeat these contents in this post but only share my experience in installing this package. Readers can start learning how to use this package from the document RAVEN mini tutorial.docx.
3.1. Installation
- Install the solver Gurobi v8.1.1 (accessed on 8 June 2019) with an academic licence.
- Set up Gurobi for MATLAB.
- Type
savepathunder the current Gurobi path in the MATLAB prompt. - Run
checkInstallationin MATLAB prompt under the RAVEN installation folder. setRavenSolver('gurobi')
Notably the authors recommend users to run RAVEN with particular KEGG and MetaCyc releases.
References
- Thiele, I. & Palsson, B. Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93 (2010).
- Karlsen, E., Schulz, C. & Almaas, E. Automated generation of genome-scale metabolic draft reconstructions based on KEGG. BMC Bioinformatics 19, 467 (2018).
- Milne, C. B. et al. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst. Biol. 5, 130 (2011).
- Martyushenko, N. & Almaas, E. ModelExplorer - software for visual inspection and inconsistency correction of genome-scale metabolic reconstructions. BMC Bioinformatics 20, 56 (2019).
- Henry, C. S. et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977 (2010).
- Metabolic Model Reconstruction, https://docs.patricbrc.org/tutorial/metabolic_model_reconstruction/metabolic_model_reconstruction.html.
- Wattam, A. R. et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591 (2013).